|
|
 | UPDATED 8/2020 |
| Augmented Annotation and Orthologue Analysis for Oryctolagus cuniculus |
|
Setting up a search:
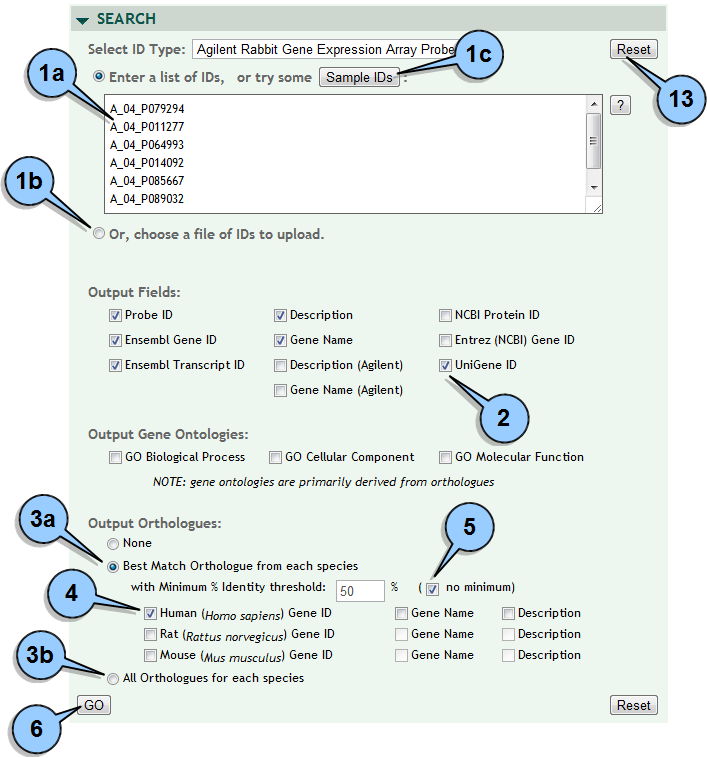
Select the type of identifier (Agilent probe, Unigene, Ensembl, Entrez, or NCBI) from the dropdown list. Enter a list of IDs by either:
(1a) typing/pasting identifiers to the text box, OR
(1b) click the radio button to choose a text file of identifiers to upload, OR
(1c) click the SAMPLE IDS button to load a short sample list of the selected type. Select the fields you want returned as part of your search by checking the appropriate "Output Fields" boxes. If you would like like to include orthologous genes in the results, then...
(3a) for only the orthologue with the highest percent identity click the radio button next to "Best Match Orthologue", OR
(3b) for all orthologous genes click the radio button next to "All Orthologues". Select one or more species (and optional gene name and/or description) for the orthologue search. To limit orthologues to only those above a certain identity threshold, uncheck "no minimum" and enter the percent threshold in the text box. Start the search by clicking the GO button. A search may take a minute or more for a large number of identifiers.
|
Evaluating search results:
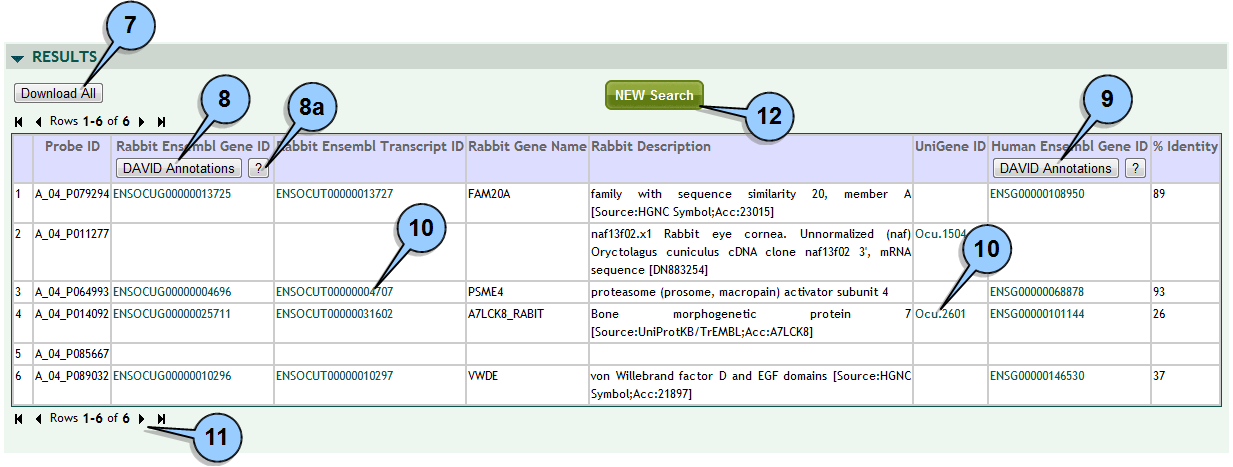
To download all search results to a file click the DOWNLOAD ALL button.
Most browsers give you the option to save the file or open it directly in a spreadsheet program. To send rabbit gene results to DAVID for functional annotation analysis click the DAVID ANNOTATIONS button at the top of the Rabbit Ensembl Gene ID column.
(8a) for step-by-step instructions on how to use DAVID, click on the '?' button.
NOTE: most rabbit genes are not functionally well annotated, so DAVID may provide little or no information. For better results try using orthologues [see Step 9]. To send orthologue gene results to DAVID for functional annotation analysis click the DAVID ANNOTATIONS button at the top of the Ensembl Gene ID column for the desired species (i.e, human, rat, mouse). To get more detailed gene information click on any gene identifier to search the corresponding external database. This is available for all Ensembl, Genbank, Entrez and Unigene identifiers.
Note that some database identifiers are for proteins not genes. Up to 50 rows are displayed per page. To skip to the start, end or move page by page, use the navigation controls at either the top or bottom of the search results pane.
|
Modifying a search:
Click on the arrow next to the SEARCH pane title to unhide the current search parameters, or click on the NEW SEARCH button in the results pane.
Modify parameters and click the GO button to rerun the search.
If identifiers are from a file you will have to re-choose the file to upload.
NOTE: depending on which browser you use, the BACK button may or may not reset the search form. Clicking on SEARCH or NEW SEARCH is preferred.
To reset the search to default parameters click the RESET button.

|
| |
|
|
|