
 |
||
|
Users' Guide
Introduction
Disulfide by Design 2 (DbD2) is a redesigned and enhanced version of our original DbD application (Dombkowski, 2003) for the rational design of disulfide bonds in proteins. For a given protein structural model, all residue pairs are rapidly assessed for proximity and geometry consistent with disulfide formation, assuming the residues were mutated to cysteines. The output displays residue pairs meeting the appropriate criteria. The input model will typically be a Protein Data Bank (PDB) structure for the protein of interest; however, structures developed through homology modeling may also be used. Engineered disulfides have proven useful for increasing the stability of proteins and to assist the investigation of protein dynamics and interactions. This software was written by Dr. Alan Dombkowski and Douglas Craig, and is based on algorithms created for disulfide identification in protein fold recognition methods (Dombkowski & Crippen, 2000). The Disulfide by Design algorithm has been successfully used for disulfide engineering in a wide variety of applications (see Papers Using DbD below). Compatibility
Disulfide by Design 2 has been tested with the following browsers:
Contact us with any compatibility issues. Getting Started
1Load Protein File
Try loading the sample PDB file by clicking on Load Example PDB
2Run Analysis
Click on the Run button at the bottom of the Options Pane. 3Evaluate Results
Inspect the analysis results. Find candidate residue pairs based on bond energy, B-factor, associated secondary structure, or 3-D considerations. 4Build Mutant Protein
Select one or more residue pairs using the checkboxes under the ANALYSIS tab. 5View & Save Results
Inspect the resulting model under the 3-D VIEWS tab. Loading PDB Files
All DbD analysis begins by loading an existing PDB file. PDB files can be loaded three different ways. 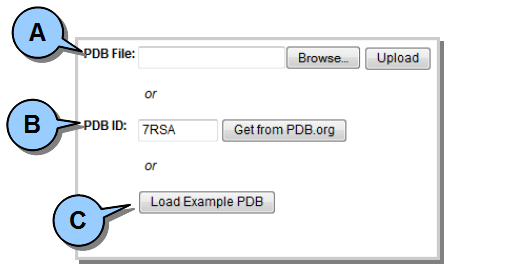 Figure 1. Loading a protein (PDB) file.
Figure 1. Loading a protein (PDB) file.
FILE INFO Tab
Once the PDB file is successfully loaded into memory, the FILE INFO tab will display summary information about the loaded protein. This information is gathered from PDB file fields. The actual content of the file will also be displayed in the scrollable/resizable Content pane. 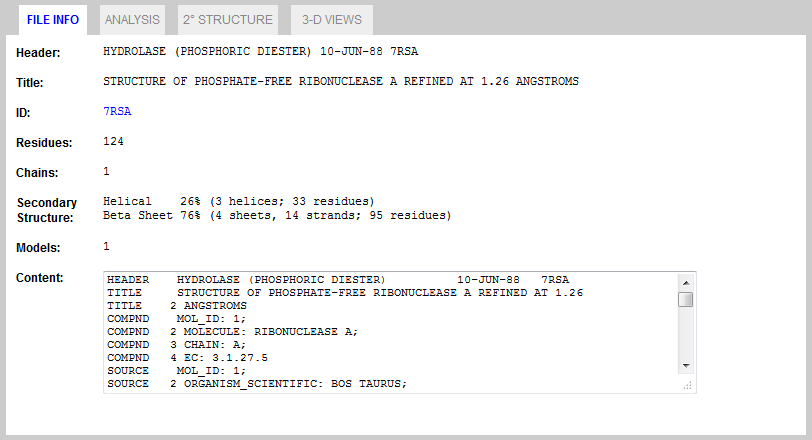 Figure 2. FILE INFO Tab.
Figure 2. FILE INFO Tab.Running Analysis
Options Pane
The Options Pane (to the left of the FILE INFO tab) is used to set all DbD analysis parameters. 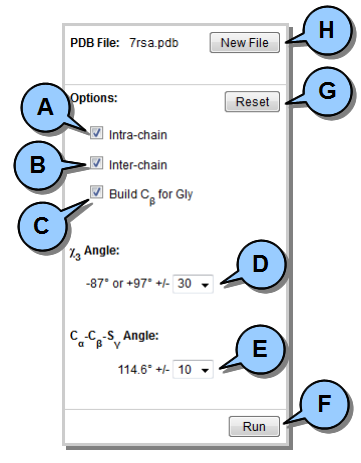 Figure 3. Options Pane.
Figure 3. Options Pane.
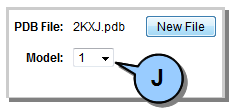 Figure 4. Multiple model selection.
Figure 4. Multiple model selection.
Evaluating Analysis Results
After running DbD analysis, three categories of information are available from which to assess results:
Each is available under a separate tab, and is described in detail below. ANALYSIS Tab
This tab displays a list of all residue pairs which analysis indicates may potentially form a disulfide bond. All information (except for energy and angle) is drawn directly from the PDB file. 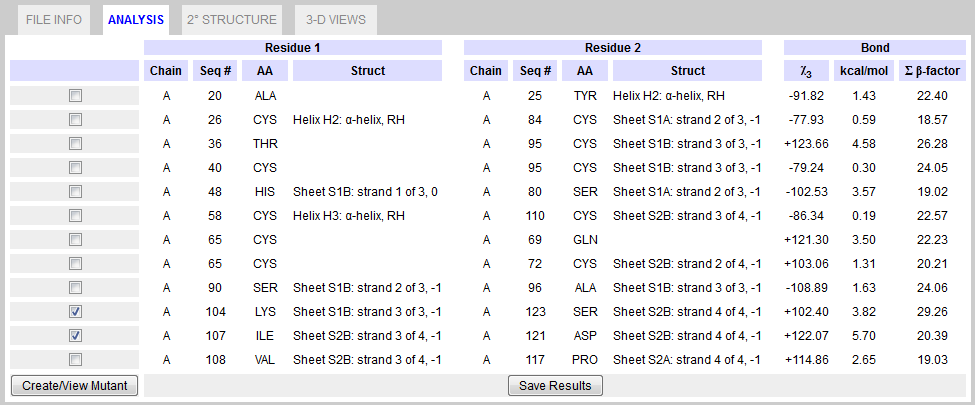 Figure 5. ANALYSIS Tab.
2° STRUCTURE Tab
This tab displays detailed secondary structure for the protein, including where potential disulfide bonds are located based on DbD analysis. All information is drawn directly from the PDB file. 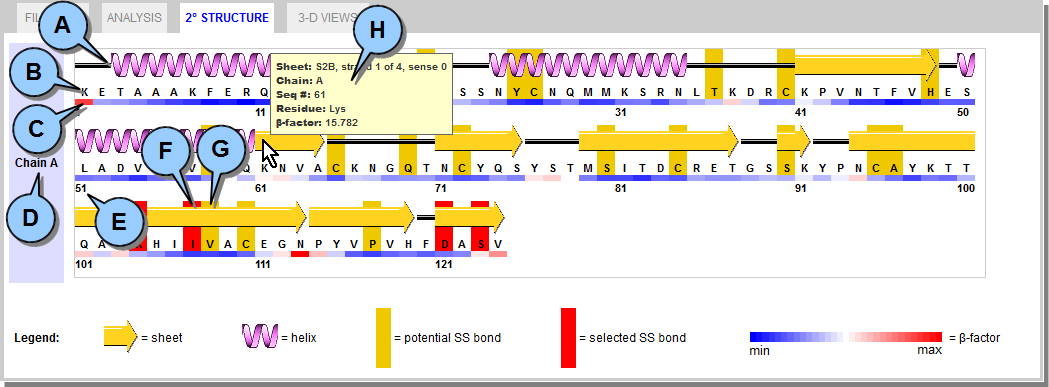 Figure 6. 2° STRUCTURE Tab.
3-D VIEWS Tab This tab displays an interactive 3D representation of the protein using the open-source Jmol Java plugin, as well as the mutated protein if one has been generated (see Building Mutant Proteins). 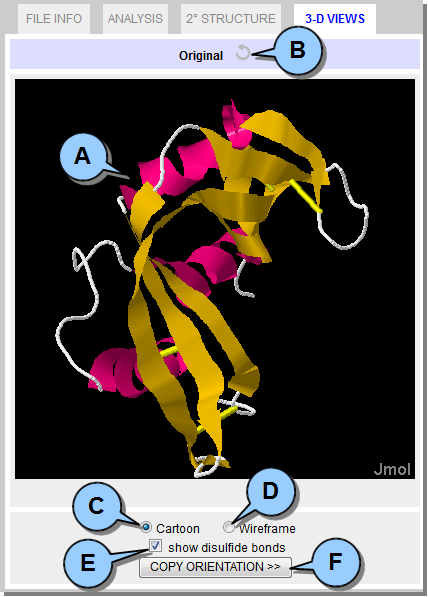 Figure 7. 3-D VIEW Tab.
Building Mutant Proteins
Once DbD analysis has been run you will have a list of residue pairs under the ANALYSIS tab which could potentially form a disulfide bond if mutated into cysteines. 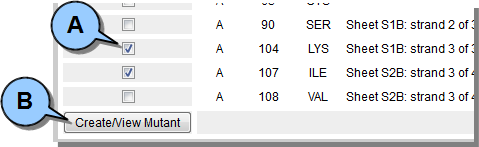 Figure 8. Select residue pairs and create mutant.
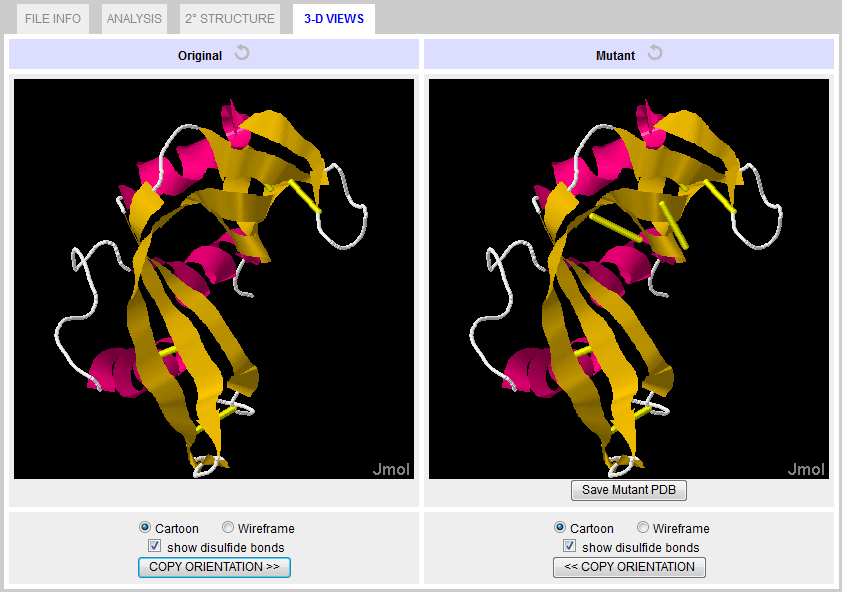 Figure 9. 3-D VIEW Tab with mutant. Saving Results & Models
The results of DbD analysis can be saved to a local text file in comma-separated value (CSV) format. 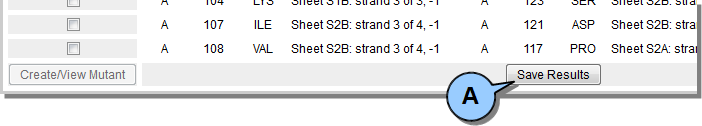 Figure 10. Save analysis results.
Figure 10. Save analysis results.
The output can then be opened in any spreadsheet application (e.g., Excel), and includes the following information: 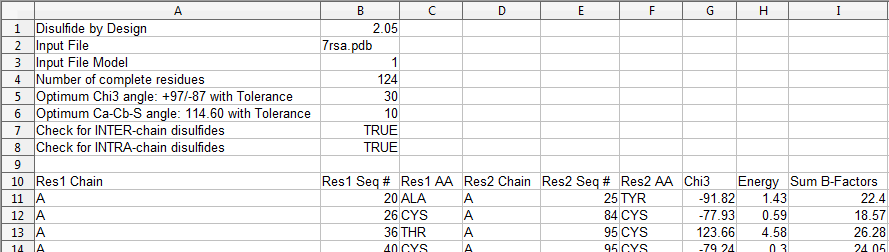 Figure 11. Analysis results CSV file contents. Any mutated protein (i.e., one with new disulfide bonds) can be saved to a new PDB file. 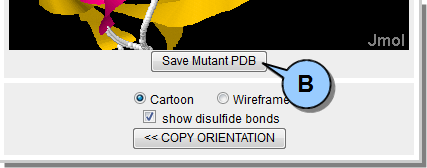 Figure 12. Save mutated PDB file.
Note that for multiple model PDB files, only the one modified model is saved to the new PDB. 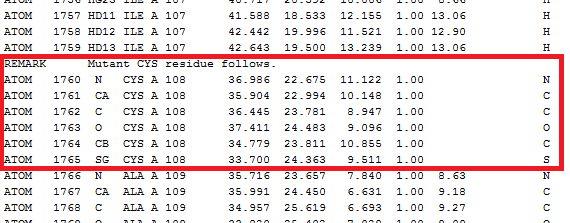 Figure 13. Mutant PDB file disulfide. Tips & Tricks
Multiple Models PDBs
When analyzing a PDB with multiple models it is possible to generate a mutant for one model of the PDB and then load a second model (using the Options Pane drop-down). This allows a side-by-side visual comparison of the two model variants under the 3-D VIEW tab. Technical Details
The Figure 14 below represents a cysteine pair coupled by a disulfide bond. The approach uses a disulfide model with fixed Cβ-Sγ and Sγ-Sγ bond lengths, 1.81 and 2.04 Ĺ respectively, and fixed Cβ-Sγ-Sγ bond angles of 104.15°. These bond lengths and angles are consistent with values observed in a survey of protein disulfide bonds (Petersen et al., 1999). The χ3 torsion angle, formed by rotation of the Cβ atoms about the Sγ-Sγ bond, is allowed to vary in the model and can be described as a function of the distance between Cβ atoms (Dombkowski & Crippen, 2000). 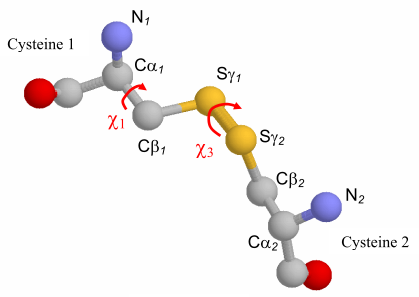 Figure 14. Anatomy of a disulfide.
Figure 14. Anatomy of a disulfide.
To "fit" the disulfide model between a pair of residues,
the algorithm simply rotates the χ3 angle to obtain a Cβ-Cβ distance that matches the Cβ-Cβ
distance measured between the residues. Numerous Sγ locations are possible, so all
possible Sγ orientations are examined and the atomic coordinates providing the lowest
energy (Eij) are selected, based on the χ1 and χ3 torsion angles and the two Cα-Cβ-Sγ
angles. The χ1 torsion angle is defined by the N-Cα-Cβ-Sγ atoms. Eij is calculated per
equations (1-4), where i and j are residue indices, θ is the Cα-Cβ-Sγ angle, and θ0 is set to
114.6°. Energy units are kcal/mol.
The energy calculation provides minima at χ3 values of +97° and -87° (Figure 15) and χ1 values of ±60° and ±180°. These values are derived from a sample of 331 proteins containing 1418 known disulfide bonds. Since the disulfide model uses fixed bond lengths, no term is included for bond stretching. 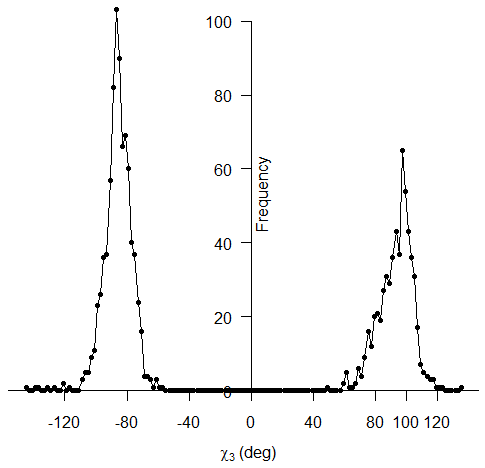 Figure 15. χ3 distribution of 1418 known disulfide bonds. The distribution of energy values for these 1418 known disulfide bonds (Figure 16) reveals a mean energy value, calculated per equations (1 -4 above), of 0.89 kcal/mol and a maximum of 8.35 kcal/mol. 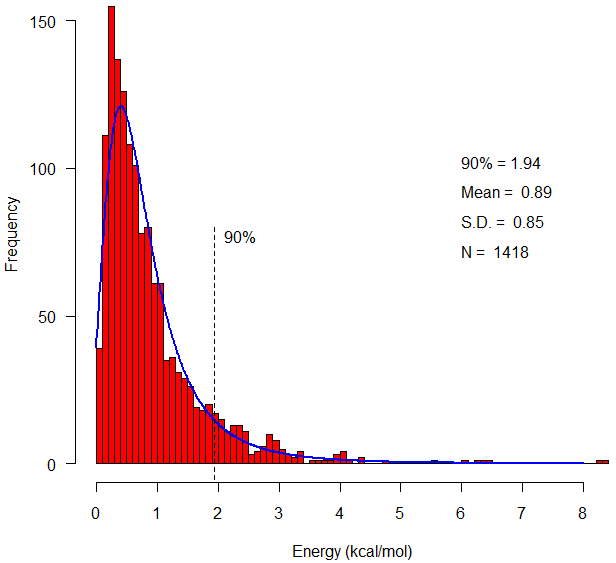 Figure 16. Energy distribution of 1418 known disulfide bonds. The energy calculation also provides a means to compare potential disulfides during disulfide design. The following Figure 17 compares the calculated energy of actual disulfide bonds to their corresponding designed disulfide bonds. 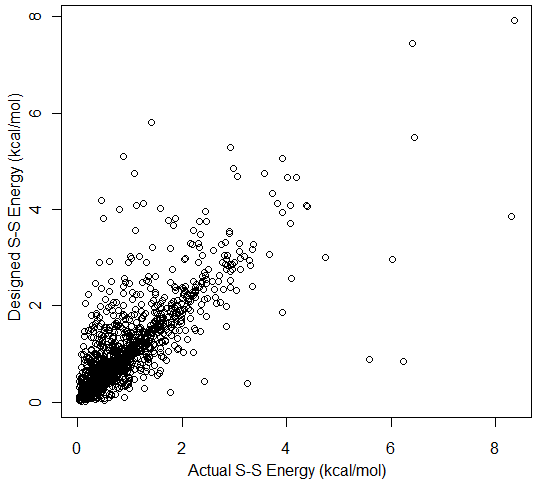 Figure 17. Energy for 1418 designed disulfides versus energy of the actual disulfides. Validation Test Set
A list of PDB identifiers used to validate DbD2 can be found here. Release Notes
Known Issues
2.12 21 May 2015
2.11 17 Sep 2013
2.10 20 Jun 2013
2.06 19 Dec 2012
2.00 1 May 2012
1.20 16 Sep 2003 (Windows Version)
1.12 1 Jan 2003 (Windows Version)
References
Craig, D.B. and Dombkowski A.A.
Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins.
BMC Bioinformatics. 2013 Dec 1;14:346.
DOI: 10.1186/1471-2105-14-346
PMID: 24289175
Dombkowski, A.A. Disulfide by Design: A computational method for the rational
design of disulfide bonds in proteins. Bioinformatics. 2003 Sep 22; 19(14):1852-3.
PMID: 14512360
Dombkowski, A.A., Sultana, K.Z., and Craig, D.B.
Protein disulfide engineering.
FEBS Letters. 2014 Jan 21;588(2):206-12.
DOI: 10.1016/j.febslet.2013.11.024
PMID: 24291258
Anthony, L.C. and Burgess, R.R. Conformational Flexibility in sigma 70 Region 2
during Transcription Initiation. J Biol Chem. 2002 Nov 29;277(48):46433-41. Epub 2002 Sep 30.
PMID: 12359719
Anthony, L.C., Dombkowski, A.A., and Burgess, R.R. Using disulfide engineering
to study conformational changes in the β’260-309 coiled-coil region of E. Coli RNA
polymerase during σ70 binding. J Bacteriol. 2002 May;184(10):2634-41.
PMID: 11976292
Dombkowski, A.A. and Crippen, G.M. Disulfide recognition in an optimized
threading potential. Protein Engineering. 2000 Oct;13(10):679-89.
PMID: 11112506
Petersen, M., Jonson, P.H., and Petersen, S.B. Amino acid neighbours and detailed
conformational analysis of cysteines in proteins. Protein Engineering. 1999 Jul;12(7):535-48.
PMID: 10436079
Papers Using the Disulfide by Design Algorithm
2018
Xie, D.F., Fang, H., Mei, J.Q., Gong, J.Y., Wang, H.P., Shen, X.Y., Huang, J., Mei, L.H.
Improving thermostability of (R)-selective amine transaminase from Aspergillus terreus through introduction of disulfide bonds.
Biotechnology and Applied Biochemistry.
2018;65(2):255-262.
DOI: 10.1002/bab.1572
PMID: 28639260
Wang, X., Lu, Z., Xu, T., Selvaraj, J.N., Yi, L., Zhang, G.
Improving the specific activity and thermo-stability of alkaline pectate lyase from Bacillus subtilis 168 for bioscouring.
Biochemical Engineering Journal.
2018;129:74-83.
DOI: 10.1016/j.bej.2017.11.001
Pandey, R.K., Ojha, R., Aathmanathan, V.S., Krishnan, M., Prajapati, V.K.
Immunoinformatics approaches to design a novel multi-epitope subunit vaccine against HIV infection.
Vaccine.
2018;36(17):2262-2272.
DOI: 10.1016/j.vaccine.2018.03.042
PMID: 29571972
Pandey, R.K., Bhatt, T.K., Prajapati, V.K.
Novel Immunoinformatics Approaches to Design Multi-epitope Subunit Vaccine for Malaria by Investigating Anopheles Salivary Protein.
Scientific Reports.
2018;8(1).
DOI: 10.1038/s41598-018-19456-1
PMID: 29348555
Li, G., Fang, X., Su, F., Chen, Y., Xu, L., Yan, Y.
Enhancing the thermostability of Rhizomucor miehei lipase with a limited screening library by rational-design point mutations and disulfide bonds.
Applied and Environmental Microbiology.
2018;84(2).
DOI: 10.1128/AEM.02129-17
PMID: 29101200
Kist, A.M., Lampert, A., O'Reilly, A.O.
DIsulfide Mapping PLanner Software Tool.
Journal of Computational Biology.
2018;25(4):430-434.
DOI: 10.1089/cmb.2017.0129
PMID: 28817312
Fass, D., Thorpe, C.
Chemistry and Enzymology of Disulfide Cross-Linking in Proteins.
Chemical Reviews.
2018;118(3):1169-1198.
DOI: 10.1021/acs.chemrev.7b00123
PMID: 28699750
Buß, O., Rudat, J., Ochsenreither, K.
FoldX as Protein Engineering Tool: Better Than Random Based Approaches?
Computational and Structural Biotechnology Journal.
2018;16:25-33.
DOI: 10.1016/j.csbj.2018.01.002
2017
Zheng, J., Yang, T., Zhou, J., Xu, M., Zhang, X., Rao, Z.
Elimination of a free cysteine by creation of a disulfide bond increases the activity and stability of Candida boidinii formate dehydrogenase.
Applied and Environmental Microbiology.
2017;83(2).
DOI: 10.1128/AEM.02624-16
PMID: 27836850
Tanghe, M., Danneels, B., Last, M., Beerens, K., Stals, I., Desmet, T.
Disulfide bridges as essential elements for the thermostability of lytic polysaccharide monooxygenase LPMO10C from Streptomyces coelicolor.
Protein Engineering, Design and Selection.
2017;30(5):401-408.
DOI: 10.1093/protein/gzx014
PMID: 28338903
Kraithong, T., Channgam, K., Itsathitphaisarn, O., Tiensuwan, M., Jeruzalmi, D., Pakotiprapha, D.
Movement of the β-hairpin in the third zinc-binding module of UvrA is required for DNA damage recognition.
DNA Repair.
2017;51:60-69.
DOI: 10.1016/j.dnarep.2017.02.003
PMID: 28209516
Khatoon, N., Pandey, R.K., Prajapati, V.K.
Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach.
Scientific Reports.
2017;7(1).
DOI: 10.1038/s41598-017-08842-w
PMID: 28811600
Hong, E.Y., Lee, S.G., Park, B.J., Lee, J.M., Yun, H., Kim, B.G.
Simultaneously Enhancing the Stability and Catalytic Activity of Multimeric Lysine Decarboxylase CadA by Engineering Interface Regions for Enzymatic Production of Cadaverine at High Concentration of Lysine.
Biotechnology Journal.
2017;12(11).
DOI: 10.1002/biot.201700278
PMID: 28843030
Hajighahramani, N., Nezafat, N., Eslami, M., Negahdaripour, M., Rahmatabadi, S.S., Ghasemi, Y.
Immunoinformatics analysis and in silico designing of a novel multi-epitope peptide vaccine against Staphylococcus aureus.
Infection, Genetics and Evolution.
2017;48:83-94.
DOI: 10.1016/j.meegid.2016.12.010
PMID: 27989662
Fattahian, Y., Riahi-Madvar, A., Mirzaee, R., Asadikaram, G., Rahbar, M.R.
In silico locating the immune-reactive segments of Lepidium draba peroxidase and designing a less immune-reactive enzyme derivative.
Computational Biology and Chemistry.
2017;70:21-30.
DOI: 10.1016/j.compbiolchem.2017.07.003
PMID: 28743101
Fanaei Kahrani, Z., Emamzadeh, R., Nazari, M., Rasa SMM.
Molecular basis of thermostability enhancement of Renilla luciferase at higher temperatures by insertion of a disulfide bridge into the structure.
Biochimica et Biophysica Acta - Proteins and Proteomics.
2017;1865(2):252-259.
DOI: 10.1016/j.bbapap.2016.11.004
PMID: 27863256
Contreras-Martel, C., Martins, A., Ecobichon, C., Trindade, D.M., Matteď, P.J., Hicham, S., Hardouin, P., Ghachi, M.E., Boneca, I.G., Dessen, A.
Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.
Nature Communications.
2017;8(1).
DOI: 10.1038/s41467-017-00783-2
PMID: 28974686
Abkallo, H.M., Martinelli, A., Inoue, M., Ramaprasad, A., Xangsayarath, P., Gitaka, J., Tang, J., Yahata, K., Zoungrana, A., Mitaka, H., Acharjee, A., Datta, P.P., Hunt, P., Carter, R., Kaneko, O., Mustonen, V., Illingworth, C.J.R., Pain, A., Culleton, R.
Rapid identification of genes controlling virulence and immunity in malaria parasites.
PLoS Pathogens.
2017;13(7).
DOI: 10.1371/journal.ppat.1006447
PMID: 28704525
2016
Yi, M.C., Khosla, C.
Thiol-Disulfide Exchange Reactions in the Mammalian Extracellular Environment.
Annual Review of Chemical and Biomolecular Engineering.
2016;7:197-222.
DOI: 10.1146/annurev-chembioeng-080615-033553
PMID: 27023663
Yagi, S., Akanuma, S., Yamagishi, M., Uchida, T., Yamagishi, A.
De novo design of protein-protein interactions through modification of inter-molecular helix-helix interface residues.
Biochimica et Biophysica Acta - Proteins and Proteomics.
2016;1864(5):479-487.
DOI: 10.1016/j.bbapap.2016.02.008
PMID: 26867971
Tan, H., Miao, R., Liu, T., Cao, X., Wu, X., Xie, L., Huang, Z., Peng, W., Gan, B.
Enhancing the thermal resistance of a novel acidobacteria-derived phytase by engineering of disulfide bridges.
Journal of Microbiology and Biotechnology.
2016;26(10):1717-1722.
DOI: 10.4014/jmb.1604.04051
PMID: 27363471
Rana, A., Akhter, Y.
A multi-subunit based, thermodynamically stable model vaccine using combined immunoinformatics and protein structure based approach.
Immunobiology.
2016;221(4):544-557.
DOI: 10.1016/j.imbio.2015.12.004
PMID: 26707618
Postel, S., Deredge, D., Bonsor, D.A., Yu, X., Diederichs, K., Helmsing, S., Vromen, A., Friedler, A., Hust, M., Egelman, E.H., Beckett, D., Wintrode, P.L., Sundberg, E.J.
Bacterial flagellar capping proteins adopt diverse oligomeric states.
eLife.
2016;5.
DOI: 10.7554/eLife.18857
PMID: 27664419
Morris, G., Fanucchi, S.
A Key Evolutionary Mutation Enhances DNA Binding of the FOXP2 Forkhead Domain.
Biochemistry.
2016;55(13):1959-1967.
DOI: 10.1021/acs.biochem.5b01271
PMID: 26950495
Mirsaeidi, M., Gidfar, S., Vu, A., Schraufnagel, D.
Annexins family: Insights into their functions and potential role in pathogenesis of sarcoidosis.
Journal of Translational Medicine.
2016;14(1).
DOI: 10.1186/s12967-016-0843-7
PMID: 27071553
Liu, T., Wang, Y., Luo, X., Li, J., Reed, S.A., Xiao, H., Young, T.S., Schultz, P.G.
Enhancing protein stability with extended disulfide bonds.
Proceedings of the National Academy of Sciences of the United States of America.
2016;113(21):5910-5915.
DOI: 10.1073/pnas.1605363113
PMID: 27162342
Kim, J.H., Song, D.H., Youn, S.J., Kim, J.W., Cho, G., Kim, S.C., Lee, H., Jin, M.S., Lee, J.O.
Crystal structures of mono- and bi-specific diabodies and reduction of their structural flexibility by introduction of disulfide bridges at the Fv interface.
Scientific Reports.
2016;6.
DOI: 10.1038/srep34515
PMID: 27682821
Jo, B.H., Park, T.Y., Park, H.J., Yeon, Y.J., Yoo, Y.J., Cha, H.J.
Engineering de novo disulfide bond in bacterial α-type carbonic anhydrase for thermostable carbon sequestration.
Scientific Reports.
2016;6.
DOI: 10.1038/srep29322
PMID: 27385052
Deller, M.C., Kong, L., Rupp, B.
Protein stability: A crystallographer's perspective.
Acta Crystallographica Section:F Structural Biology Communications.
2016;72:72-95.
DOI: 10.1107/S2053230X15024619
PMID: 26841758
Chino, M., Leone, L., Maglio, O., Lombardi, A.
Designing Covalently Linked Heterodimeric Four-Helix Bundles.
Methods in Enzymology.
2016;580:471-499.
DOI: 10.1016/bs.mie.2016.05.036
PMID: 27586346
Bergal, H.T., Hopkins, A.H., Metzner, S.I., Sousa, M.C.
The Structure of a BamA-BamD Fusion Illuminates the Architecture of the β-Barrel Assembly Machine Core.
Structure.
2016;24(2):243-251.
DOI: 10.1016/j.str.2015.10.030
PMID: 26749448
2015
Wołek, K., Gómez-Sicilia, Ŕ., Cieplak, M.
Determination of contact maps in proteins: A combination of structural and chemical approaches.
Journal of Chemical Physics.
2015;143(24).
DOI: 10.1063/1.4929599
PMID: 26723590
Vinther, T.N., Pettersson, I., Huus, K., Schlein, M., Steensgaard, D.B., Sřrensen, A., Jensen, K.J., Kjeldsen, T., Hubalek, F.
Additional disulfide bonds in insulin: Prediction, recombinant expression, receptor binding affinity, and stability.
Protein Science.
2015;24(5):779-788.
DOI: 10.1002/pro.2649
PMID: 25627966
Fogen, D., Wu, S.C., Ng, K.K.S., Wong, S.L.
Engineering streptavidin and a streptavidin- binding peptide with infinite binding affinity and reversible binding capability: Purification of a tagged recombinant protein to high purity via affinity-driven thiol coupling.
PLoS ONE.
2015;10(9).
DOI: 10.1371/journal.pone.0139137
PMID: 26406477
Yin, X., Hu, D., Li, J. F., He, Y., Zhu, T. D., Wu, M. C.
Contribution of Disulfide Bridges to the Thermostability of a Type A Feruloyl Esterase from Aspergillus usamii.
PLoS One. 2015 May 13;10(5):e0126864.
DOI: 10.1371/journal.pone.0126864
PMID: 25969986
Bonsor, D. A., Pham, K. T., Beadenkopf, R., Diederichs, K., Haas, R., Beckett, D., Fischer, W., Sundberg, E. J.
Integrin engagement by the helical RGD motif of the Helicobacter pylori CagL protein is regulated by pH-induced displacement of a neighboring helix.
The Journal of Biological Chemistry. 2015 Apr 2. pii: jbc.M115.641829.
PMID: 25837254
Dupré, E., Herrou, J., Lensink, M. F., Wintjens, R., Vagin, A., Lebedev, A., Crosson, S., Villeret, V., Locht, C., Antoine, R., Jacob-Dubuisson, F.
Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS.
PLoS Pathogens. 2015 Mar 4;11(3):e1004700.
DOI: 10.1371/journal.ppat.1004700
PMID: 25738876
Jansen, K.B., Baker, S.L., Sousa, M.C.
Crystal structure of BamB bound to a periplasmic domain fragment of BamA, the central component of the β-barrel assembly machine.
The Journal of Biological Chemistry. 2015 Jan 23;290(4):2126-36.
DOI: 10.1074/jbc.M114.584524
PMID: 25468906
Touw, W.G., Baakman, C., Black, J., te Beek, T. A., Krieger, E., Joosten, R. P., Vriend, G.
A series of PDB-related databanks for everyday needs.
Nucleic Acids Research. 2015 Jan;43(Database issue):D364-8.
DOI: 10.1093/nar/gku1028
PMID: 25352545
2014
Quistgaard, E. M.
A disulfide polymerized protein crystal.
Chemical Communications. 2014 Dec 11;50(95):14995-7.
DOI: 10.1039/c4cc07326f
PMID: 25327138
Zhang, S., Wang, Y., Song, X., Hong, J., Zhang, Y., Yao, L.
Improving Trichoderma reesei Cel7B Thermostability by Targeting the Weak Spots.
Journal of Chemical Information and Modeling. 2014 Oct 6. [Epub ahead of print].
PMID: 25286389
Roderer, D., Benke, S., Müller, M., Fäh-Rechsteiner, H., Ban, N., Schuler, B., Glockshuber, R.
Characterization of Variants of the Pore-Forming Toxin ClyA from Escherichia coli Controlled by a Redox Switch.
Biochemistry. 2014 Sep 30; [Epub ahead of print].
PMID: 25222267
Tan, Z., Li, J., Wu, M., Wang, J.
Enhancing the Thermostability of a Cold-Active Lipase from Penicillium cyclopium by In Silico Design of a Disulfide Bridge.
Applied Biochemistry and Biotechnology. 2014 May 28; [Epub ahead of print].
PMID: 24867629
Surzhik, M. A. , Schmidt, A. E. , Glazunov, E. A. , Firsov, D. L. , Petukhov, M. G.
Introduction of additional thiol groups into glucoamylase in Aspergillus Awamori and their effect on the thermal stability and catalytic activity of the enzyme.
Applied Biochemistry and Microbiology. 2014 Mar;50(2):118-124.
Sanchez, J. G. , Okreglicka, K. , Chandrasekaran, V. , Welker, J. M. , Sundquist, W. I. , Pornillos, O.
The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer.
PNAS. 2014 ; published ahead of print February 3, 2014.
DOI: 10.1073/pnas.1318962111
PMID: 24550273
Liu, L., Deng, Z., Yang, H., Li, J., Shin, H.D., Chen, R.R., Du, G., Chen, J.
In silico rational design and systems engineering of disulfide bridges in the catalytic domain of an alkaline α-amylase from Alkalimonas amylolytica to improve thermostability.
Appl Environ Microbiol. 2014 Feb;80(3):798-807.
DOI: 10.1128/AEM.03045-13
PMID: 24212581
Kim, D.Y., To, R., Kandalaft, H., Ding, W., van Faassen, H., Luo, Y., Schrag, J.D., St-Amant, N., Hefford, M., Hirama, T., Kelly, J.F., MacKenzie, R., Tanha, J.
Antibody light chain variable domains and their biophysically improved versions for human immunotherapy.
MAbs. 2014 Jan-Feb;6(1):219-35.
DOI: 10.4161/mabs.26844
PMID: 24423624
2013
Shinwari, J. , Tahir, A. , Bohlega, S. and AlTassan, N.
In silico analysis of influence of the missense mutation P629S on the molecular interaction and 3D properties of PIK3R5.
Advances in Biological Chemistry. 2013; 3:408-417.
DOI: 10.4236/abc.2013.34044
Housden, N.G., Hopper, J.T., Lukoyanova, N., Rodriguez-Larrea, D., Wojdyla, J.A., Klein, A., Kaminska, R., Bayley, H., Saibil, H.R., Robinson, C.V., Kleanthous, C.
Intrinsically disordered protein threads through the bacterial outer-membrane porin OmpF.
Science. 2013 Jun 28;340(6140):1570-4.
DOI: 10.1126/science.1237864
PMID: 23812713
Gosein, V., Miller, G.J.
Conformational Stability of Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase (IPK1) Dictates Its Substrate Selectivity.
J Biol Chem. 2013 Dec 27;288(52):36788-95. Epub 2013 Oct 28.
DOI: 10.1074/jbc.M113.512731
PMID: 24165122
Geng, Y., Bush, M., Mosyak, L., Wang, F., Fan, Q.R.
Structural mechanism of ligand activation in human GABAB receptor.
Nature. 2013 Dec 4. [Epub ahead of print].
DOI: 10.1038/nature12725
PMID: 24305054
Ding, H., Gao, F., Liu, D., Li, Z., Xu, X., Wu, M., Zhao, Y.
Significant improvement of thermal stability of glucose 1-dehydrogenase by introducing disulfide bonds at the tetramer interface.
Enzyme Microb Technol. 2013 Dec 10;53(6-7):365-72. Epub 2013 Aug 16.
DOI: 10.1016/j.enzmictec.2013.08.001
PMID: 24315638
Ziarek, J.J., Getschman, A.E., Butler, S.J., Taleski, D., Stephens, B., Kufareva, I., Handel, T.M., Payne ,R.J., Volkman, B.F.
Sulfopeptide probes of the CXCR4/CXCL12 interface reveal oligomer-specific contacts and chemokine allostery.
ACS Chem Biol. 2013 Sep 20;8(9):1955-63. Epub 2013 Jun 26.
DOI: 10.1021/cb400274z
PMID: 23802178
Housden, N.G., Hopper, J.T., Lukoyanova, N., Rodriguez-Larrea, D., Wojdyla, J.A., Klein, A., Kaminska, R., Bayley, H., Saibil, H.R., Robinson, C.V., Kleanthous, C.
Intrinsically disordered protein threads through the bacterial outer-membrane porin OmpF.
Science. 2013 Jun 28;340(6140):1570-4.
DOI: 10.1126/science.1237864
Farrance, O.E., Hann, E., Kaminska, R., Housden, N.G., Derrington, S.R., Kleanthous, C., Radford, S.E., Brockwell, D.J.
A Force-Activated Trip Switch Triggers Rapid Dissociation of a Colicin from Its Immunity Protein.
PLoS Biol. 2013 Feb 19; 11(2): e1001489.
DOI: 10.1371/journal.pbio.1001489
McConnell, A.D., Spasojevich, V., Macomber, J.L., Krapf, I.P., Chen, A., Sheffer, J.C., Berkebile, A., Horlick, R.A., Neben, S., King, D.J., Bowers, P.M.
An integrated approach to extreme thermostabilization and affinity maturation of an antibody.
Protein Engineering, Design and Selection. 2013 Feb;26(2):151-163.
PMID: 23173178
2012
Hoffmann, A., Becker, A.H., Zachmann-Brand, B., Deuerling, E., Bukau, B., Kramer, G.
Concerted action of the ribosome and the associated chaperone trigger factor confines nascent polypeptide folding.
Molecular Cell. 2012 Oct 12;48(1):63-74.
DOI: 10.1016/j.molcel.2012.07.018
PMID: 22921937
Doreleijers, J.F., Sousa da Silva, A.W., Krieger, E., Nabuurs, S.B., Spronk, C.A., Stevens, T.J., Vranken, W.F., Vriend, G., Vuister, G.W.
CING: an integrated residue-based structure validation program suite..
Journal of Biomolecular NMR. 2012 Nov;54(3):267-83.
DOI: 10.1007/s10858-012-9669-7
PMID: 22986687
Saman Hosseinkhani, Mahboobeh Nazari and Leila Hassani.
Step-wise addition of disulfide bridge in firefly luciferase controls color shift through a flexible loop: A thermodynamic perspective.
Photochemical & Photobiological Sciences. 2012 Sep 04.
DOI: 10.1039/C2PP25140J
Kim, D.Y., Kandalaft, H., Ding, W., Ryan, S., van Faassen, H., Hirama, T., Foote, S.J., Mackenzie, R., Tanha, J.
Disulfide linkage engineering for improving biophysical properties of human VH domains.
Protein Engineering, Design & Selection. 2012 Aug 30. [Epub ahead of print].
PMID: 22942392
Glynn, S.E., Nager, A.R., Baker, T.A., Sauer, R.T.
Dynamic and static components power unfolding in topologically closed rings of a AAA+ proteolytic machine.
Nature Structural and Molecular Biology. 2012 May 6; 19(6):616-622.
PMID: 22562135
Le, Q.A.T., Joo, J.C., Yoo, Y.J., Kim, Y.H.
Development of thermostable Candida antarctica lipase B through novel in silico design of disulfide bridge.
Biotechnology and Bioengineering. 2012 Apr; 109(4):867-876.
PMID: 22095554
Deaconescu, A.M., Sevostyanova, A., Artsimovitch, I., Grigorieff, N.
Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface.
Proc Natl Acad Sci U S A. 2012 Feb 28; 109(9):3353-8.
PMID: 22331906
[see Supplemental Information]
Badieyan, S., Bevan, D.R., Zhang, C.
Study and design of stability in GH5 cellulases.
Biotechnology and Bioengineering. 2012 Jan; 109(1):31-44.
PMID: 21809329
2011
Härd, T.
Protein engineering to stabilize soluble amyloid β-protein aggregates for structural and functional studies.
FEBS Journal. 2011 Oct; 278(20):3884-3892.
PMID: 21824290
Nazari, M., Hosseinkhani, S.
Design of disulfide bridge as an alternative mechanism for color shift in firefly luciferase and development of secreted luciferase.
Photochemical and Photobiological Sciences. 2011 Jul; 10(7):1203-1215.
PMID: 21494742
Compton, J.R., Legler, P.M., Clingan, B.V., Olson, M.A., Millard, C.B.
Introduction of a disulfide bond leads to stabilization and crystallization of a ricin immunogen.
Proteins: Structure, Function and Bioinformatics. 2011 Apr; 79(4):1048-1060.
PMID: 21387408
Huang, C.-C., Orban, T., Jastrzebska, B., Palczewski, K., Tesmer, J.J.G.
Activation of G protein-coupled receptor kinase 1 involves interactions between its N-terminal region and its kinase domain.
Biochemistry. 2011 Mar 22; 50(11):1940-1949.
PMID: 21265573
2010
Muskotál A, Seregélyes C, Sebestyén A, Vonderviszt F.
Structural basis for stabilization of the hypervariable D3 domain of Salmonella flagellin upon filament formation.
J Mol Biol. 2010 Nov 5;403(4):607-15. Epub 2010 Sep 22.
PMID: 20868693
Han L, Monné M, Okumura H, Schwend T, Cherry AL, Flot D, Matsuda T, Jovine L.
Insights into egg coat assembly and egg-sperm interaction from the X-ray structure of full-length ZP3.
Cell. . 2010 Oct 29;143(3):404-15. Epub 2010 Oct 21.
PMID: 20970175
[see Supplemental Information]
Logan T, Clark L, Ray SS.
Engineered disulfide bonds restore chaperone-like function of DJ-1 mutants linked to familial Parkinson's disease.
Biochemistry. 2010 Jul 13;49(27):5624-33.
PMID: 20527929
Xingzhou Chen, Shunqing Xu, Maosheng Zhu, Luosheng Cui, Hui Zhu, Yunxiang Liang, Zhongming Zhang
Site-directed mutagenesis of an Aspergillus niger xylanase B and its expression, purification and enzymatic characterization in Pichia pastoris.
Process Biochemistry. Volume 45, Issue 1, January 2010, Pages 75-80.
DOI: 10.1016/j.procbio.2009.08.009 2009
Wu SC, Ng KK, Wong SL.
Engineering monomeric streptavidin and its ligands with infinite affinity in binding but reversibility in interaction.
Proteins. 2009 Nov 1;77(2):404-12.
PMID: 19425108
Han ZL, Han SY, Zheng SP, Lin Y.
Enhancing thermostability of a Rhizomucor miehei lipase by engineering a disulfide bond and displaying on the yeast cell surface.
Appl Microbiol Biotechnol. 2009 Nov;85(1):117-26. Epub 2009 Jun 17.
PMID: 19533118
Zhang H, Kenaan C, Hamdane D, Hoa GH, Hollenberg PF.
Effect of conformational dynamics on substrate recognition and specificity as probed by the introduction of a de novo disulfide bond into cytochrome P450 2B1.
J Biol Chem. 2009 Sep 18;284(38):25678-86. Epub 2009 Jul 15.
PMID: 19605359
Bornschlögl T, Anstrom DM, Mey E, Dzubiella J, Rief M, Forest KT.
Tightening the knot in phytochrome by single-molecule atomic force microscopy.
Biophys J. 2009 Feb 18;96(4):1508-14.
PMID: 19217867
Jankun J, Aleem AM, Selman SH, Basrur V, Skrzypczak-Jankun E.
VLHL plasminogen activator inhibitor spontaneously reactivates from the latent to active form.
Int J Mol Med. 2009 Jan;23(1):57-63.
PMID: 19082507
2008
Mateo R, Luna E, Rincón V, Mateu MG.
Engineering viable foot-and-mouth disease viruses with increased thermostability as a step in the development of improved vaccines.
J Virol. 2008 Dec;82(24):12232-40. Epub 2008 Oct 1.
PMID: 18829763
Guo Q, Jureller JE, Warren JT, Solomaha E, Florián J, Tang WJ.
Protein-protein docking and analysis reveal that two homologous bacterialadenylyl cyclase toxins interact with calmodulin differently.
J Biol Chem. 2008 Aug 29;283(35):23836-45.
PMID: 18583346
Seeger MA, von Ballmoos C, Eicher T, Brandstätter L, Verrey F, Diederichs K, Pos KM.
Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB.
Nat Struct Mol Biol. 2008 Feb;15(2):199-205. Epub 2008 Jan 27.
PMID: 18223659
2007
Sjoelund V and Kaltashov IA.
Transporter-to-Trap Conversion: a Disulfide Bond Formation in
Cellular Retinoic Acid Binding Protein I Mutant Triggered by
Retinoic Acid Binding Irreversibly Locks the Ligand Inside the
Protein.
Biochemistry. 2007 Nov 20;46(46):13382-90.
PMID: 17958379
Dubey VK, Lee J, Somasundaram T, Blaber S, Blaber M.
Spackling the
crack: stabilizing human fibroblast growth factor-1 by targeting the
N and C terminus β-strand interactions.
J Mol Biol. 2007 Aug
3;371(1):256-68. Epub 2007 May 31. PMID: 17570396
Asgeirsson B, Adalbjörnsson BV, Gylfason GA.
Engineered disulfide bonds increase active-site local stability and reduce catalytic activity of a cold-adapted alkaline phosphatase.
Biochim Biophys Acta. 2007 Jun;1774(6):679-87. Epub 2007 Apr 5.
PMID: 17493882
Zloh M, Shaunak S, Balan S, Brocchini S.
Identification and
insertion of 3-carbon bridges in protein disulfide bonds: a
computational approach.
Nat Protoc. 2007;2(5):1070-83.
PMID: 17545999
2006
Brocchini S, Balan S, Godwin A, Choi
JW, Zloh M, Shaunak S.
PEGylation of native disulfide bonds in
proteins.
Nature Protocols. 2006;1(5):2241-52. PMID: 17406463
Shen Y,
Joachimiak A, Rosner MR, Tang WJ.
Structures of human
insulin-degrading enzyme reveal a new substrate recognition
mechanism.
Nature. 2006 Oct 19;443(7113):870-4. Epub 2006 Oct 11.
PMID: 17051221
Pellequer JL, Chen SW.
Multi-template approach to modeling
engineered disulfide bonds.
Proteins. 2006 Oct 1;65(1):192-202. PMID:
16807887
Bhushan S, Johnson KA, Eneqvist T, Glaser E.
Proteolytic
mechanism of a novel mitochondrial and chloroplastic PreP
peptidasome.
Biol Chem. 2006 Aug;387(8):1087-90. PMID: 16895479
Meinhold D, Beach M, Shao Y, Osuna R, Colon W.
The location of an
engineered inter-subunit disulfide bond in factor for inversion
stimulation (FIS) affects the denaturation pathway and cooperativity.
Biochemistry. 2006 Aug 15;45(32):9767-77. PMID: 16893178
Cui C, Zhao W, Chen J, Wang J, Li Q.
Elimination of in vivo
cleavage between target protein and intein in the intein-mediated
protein purification systems.
Protein Expr Purif. 2006
Nov;50(1):74-81. Epub 2006 Jun 15. PMID: 16884922
Jeong MY, Kim S, Yun CW, Choi YJ, Cho SG.
Engineering a de novo
internal disulfide bridge to improve the thermal stability of
xylanase from Bacillus stearothermophilus No. 236.
J Biotechnol.
2006 Jul 16. PMID: 16919348
Jai Kartik V, Lavanya T, Guruprasad K.
Analysis of disulphide bond connectivity patterns in protein tertiary structure.
Int J Biol Macromol. 2006 May 30;38(3-5):174-9. Epub 2006 Apr 3.
PMID: 16580722
Laptenko O, Kim SS, Lee J, Starodubtseva M, Cava F, Berenguer J,
Kong XP,Borukhov S.
pH-dependent conformational switch activates the
inhibitor of transcription elongation.
EMBO J. 2006 May
17;25(10):2131-41. Epub 2006 Apr 20. PMID: 16628221
Johnson KA, Bhushan S, Stahl A, Hallberg BM, Frohn A, Glaser E,
Eneqvist T.
The closed structure of presequence protease PreP forms
a unique 10,000 Å3 chamber for proteolysis.
EMBO J. 2006 May
3;25(9):1977-86. Epub 2006 Apr 6. PMID: 16601675
2005
Ma K, Temiakov D, Anikin M, McAllister WT.
Probing conformational
changes in T7 RNA polymerase during initiation and termination by
using engineered disulfide linkages.
Proc Natl Acad Sci U S A. 2005
Dec 6;102(49):17612-7. Epub 2005 Nov 21. PMID: 16301518
Karlsson M, Martensson LG, Karlsson C, Carlsson U.
Denaturant-assisted formation of a stabilizing disulfide bridge from
engineered cysteines in nonideal conformations.
Biochemistry. 2005
Mar 8;44(9):3487-93. PMID: 15736958
2002
Anthony LC, Burgess RR.
Conformational flexibility in sigma70
region 2 during transcription initiation.
J Biol Chem. 2002 Nov
29;277(48):46433-41. Epub 2002 Sep 30. PMID: 12359719
Anthony LC, Dombkowski AA, Burgess RR.
Using disulfide bond
engineering to study conformational changes in the β'260-309
coiled-coil region of Escherichia coli RNA polymerase during
σ70 binding.
J Bacteriol. 2002 May;184(10):2634-41. PMID:
11976292
|